PACKAGE = ccdred TASK = ccdproc images = ccd*.imh List of CCD images to correct (ccdtype= ) CCD image type to correct (max_cac= 0) Maximum image caching memory (in Mbytes) (noproc = no) List processing steps only? (fixpix = no) Fix bad CCD lines and columns? (oversca= yes) Apply overscan strip correction? (trim = yes) Trim the image? (zerocor= no) Apply zero level correction? (darkcor= no) Apply dark count correction? (flatcor= no) Apply flat field correction? (illumco= no) Apply illumination correction? (fringec= no) Apply fringe correction? (readcor= no) Convert zero level image to readout correction? (scancor= no) Convert flat field image to scan correction? (readaxi= line) Read out axis (column|line) (fixfile= ) File describing the bad lines and columns (biassec= [2033:2047,*]) Overscan strip image section (trimsec= [120:1940,*]) Trim data section (zero = ) Zero level calibration image (dark = ) Dark count calibration image (flat = ) Flat field images (illum = ) Illumination correction images (fringe = ) Fringe correction images (minrepl= 1.) Minimum flat field value (scantyp= shortscan) Scan type (shortscan|longscan) (nscan = 1) Number of short scan lines (interac= no) Fit overscan interactively? (functio= chebyshev) Fitting function (order = 1) Number of polynomial terms or spline pieces (sample = *) Sample points to fit (naverag= -100) Number of sample points to combine (niterat= 1) Number of rejection iterations (low_rej= 3.) Low sigma rejection factor (high_re= 3.) High sigma rejection factor (grow = 0.) Rejection growing radius (mode = ql)
Imtransp ccd*.imh[*,-*] transp*.imh
PACKAGE = longslit
TASK = transform
input = "@inp" Input images
output = "@out" Output images
fitnames = "cosmicdist" Names of coordinate fits in the database
(database = "database") Identify database
(interptype = "spline3") Interpolation type
(x1 = INDEF) Output starting x coordinate
(x2 = INDEF) Output ending x coordinate
(dx = INDEF) Output X pixel interval
(nx = INDEF) Number of output x pixels
(xlog = no) Logarithmic x coordinate?
(y1 = INDEF) Output starting y coordinate
(y2 = INDEF) Output ending y coordinate
(dy = INDEF) Output Y pixel interval
(ny = INDEF) Number of output y pixels
(ylog = no) Logarithmic y coordinate?
(flux = no) Conserve flux per pixel?
(logfiles = "STDOUT,logfile") List of log files
(mode = "ql")
I call the output transf*.imh
Imtransp transf*.imh[*,-*] rot*.imh
PACKAGE = immatch
TASK = geotran
input = "@out4" Input data
output = "@out3" Output data
database = "apinmap2.map" Name of geomap database file
transforms = "@transf.in" Names of coordinate transforms in database file
(geometry = "geometric") Transformation type (linear,geometric)
(xin = INDEF) X origin of input image in pixels
(yin = INDEF) Y origin of input image in pixels
(xshift = INDEF) X origin shift in pixels
(yshift = INDEF) Y origin shift in pixels
(xout = INDEF) X origin of output image in reference units
(yout = INDEF) Y origin of output image in reference units
(xmag = INDEF) X scale of input image in pixels per reference
(ymag = INDEF) Y scale of input image in pixels per reference
(xrotation = INDEF) X rotation angle in degrees
(yrotation = INDEF) Y rotation angle in degrees
(xmin = INDEF) Minimum reference x value of output image
(xmax = INDEF) Maximum reference x value of output image
(ymin = INDEF) Minimum reference y value of output image
(ymax = INDEF) Maximum reference y value of output image
(xscale = 1.) X scale of output image in reference units per
(yscale = 1.) Y scale of output image in reference units per
(ncols = 1789) Number of columns in the output image
(nlines = 2048) Number of lines in the output image
(xsample = 1.) Coordinate surface sampling interval in x
(ysample = 1.) Coordinate surface sampling interval in y
(interpolant = "linear") Interpolant (nearest,linear,poly3,poly5,spline3
(boundary = "nearest") Boundary extension (nearest,constant,reflect,wr
(constant = 0.) Constant for constant boundary extension
(fluxconserve = yes) Preserve image flux ?
(nxblock = 512) X dimension of working block size in pixels
(nyblock = 512) Y dimension of working block size in pixels
(verbose = yes) Print messages about the progress of the task ?
(mode = "ql")
Output is geo*.imh
Imtransp geo*.imh[*,-*] georot*.imh
[*,4:47] 1
[*,59:99] 2
etc.
Set xzap parameters as:
inlist = "rot117.1" Image(s) for cosmic ray cleaning
outlist = "foo.1" Output image(s)
(zboxsz = 3) Box size for zapping
(nsigma = 4.) Number of sky sigma for zapping threshold
(nnegsigma = 5.) Number of sky sigma for negative zapping
(nrings = 1) Number of pixels to flag as buffer around
(nobjsigma = 0.) Number of sky sigma for object identifica
(skyfiltsize = 1) Median filter size for local sky evalua
(skysubsample = 1) Block averaging factor before median fi
(ngrowobj = 0) Number of pixels to flag as buffer arou
(statsec = "") Image section to use for computing sky si
(deletemask = no) Delete CR mask after execution?
(cleanpl = yes) Delete other working .pl masks after exec
(cleanimh = yes) Delete working .imh images after executio
(verbose = yes) Verbose output?
(unzap = no) Unzap using objects from OBJmask files?
(checklimits = yes) Check min and max pix values before filte
(zmin = 0.) Minimum data value for fmedian
(zmax = 32767.) Minimum data value for fmedian
(inimglist = "")
(outimglist = "")
(statlist = "")
(mode = "ql")
Set background parameters:
input = "rot118.1" Input images to be background subtracted
output = "foo1" Output background subtracted images
(axis = 1) Axis along which background is fit and subtract
(interactive = yes) Set fitting parameters interactively?
(sample = "*") Sample of points to use in fit
(naverage = -5) Number of points in sample averaging
(function = "chebyshev") Fitting function
(order = 1) Order of fitting function
(low_reject = 1.5) Low rejection in sigma of fit
(high_reject = 1.5) High rejection in sigma of fit
(niterate = 1) Number of rejection iterations
(grow = 1.) Rejection growing radius
(graphics = "stdgraph") Graphics output device
(cursor = "") Graphics cursor input
(mode = "ql")
PACKAGE = ccdred
TASK = slitredux
imprefix = "georot" Root ID for initial input images
outroot = "proc" Root for output images and files
(images = "117") Numbers of science images
(flats = "134,135,136") Numbers of flat images
(arcs = "119") Numbers of arc images
(showmode = no) Show processing steps only?
(secsfile = "475.1a.1.secs") File with image sections and IDs, def=outroot.
(offsets = "proc.offsets") File with image xy offsets, def=outroot.offsets
(rotprefix = "rot") Root ID for rotated images
(szprefix = "sz") Root ID for szapped images
(bgprefix = "bg") Root ID for sky subtracted images
(flat = "flat") Name of flat
(rflat = "rflat") Name of response-corrected flat
(do_ccdproc = no) Do ccdproc stage?
(numamps = 1) Number of amps used in readout
(biassec = "[*,2032:2047]") Bias section for one amp ccdproc
(trimsec = "[*,130:1918]") Trim section for one amp ccdproc
(gainrat = 1.0695) ratio of left gain to right, for two amp ccdpro
(do_flatsum = yes) Do flatsum stage?
(do_response = yes) Do response stage?
(sample = "10:2038") Sampling x range for response fitting
(func = "spline3") Function for fitting response
(order = 30) Order for response function fitting
(dispaxis = 1) Dispersion axis
(do_flatfield = yes) Do flatfield stage?
(do_cutup = yes) Do cutup stage?
(do_rotate = yes) Do rotate stage?
(delete_in = yes) Delete old images after rotation?
(do_szap = yes Do szap stage?
(szinter = no) Interactive szapping?
(szorder = 4) Order for background fitting?
(szlow_rej = 4.) N sigma for low rejection threshold
(szhigh_rej = 4.) N sigma for high rejection threshold
(szniter = 3) Number of iterations for bad pixel rejection
(szgrow = 1) Grow radius for bad pixel rejection
(sznavg = -5) Naverage for background fitting
(szboxsz = 5) Zapping box size
(sznsigma = 5.) Number of sigma for positive zapping threshold
(sznnegsigma = 5.) Number of sigma for negative zapping threshold
(sznrings = 1) Growth radius around cosmic rays
(do_bkgd = yes) Do bkgd stage?
(bginter = no) Interactive background fitting?
(bgfunction = "chebyshev") Fitting function
(bgaxis = 1) Axis along which to fit & subtract sky
(fitorder = 2) Order for background fitting
(bglow_rej = 1.) N sigma for low rejection threshold
(bghigh_rej = 1.4) N sigma for high rejection threshold
(bgniter = 2) Number of iterations for bad pixel rejection
(bggrow = 1) Grow radius for bad pixel rejection
(bgnavg = -3) Naverage for background fitting
(do_bpm = yes) Do bpm stage?
(do_sum = yes) Sum science images?
(do_arcsum = yes) Sum arc image(s) correspondingly?
(arcname = "") Name for summed arc image, def=outrootarc
(expname = "elaptime") Header keyword for exposure times
(do_smosaic = no) Do smosaic stage?
(tvfile = "") Name of tvmark file to make, def=outroot.tvmark
(reverse = no) Reverse direction of mosaic?
(ninter = 0) Interval to skip (in pixels) between spectra
(blankval = -50.) Value for filling blank pixels
(ifile = "")
(ifile2 = "")
(mode = "ql")
PACKAGE = apextract
TASK = apall
input = @proclist List of input images
(output = @speclist) List of output spectra
(apertur= ) Apertures
(format = multispec) Extracted spectra format
(referen= ) List of aperture reference images
(profile= ) List of aperture profile images
(interac= yes) Run task interactively?
(find = yes) Find apertures?
(recente= yes) Recenter apertures?
(resize = yes) Resize apertures?
(edit = yes) Edit apertures?
(trace = yes) Trace apertures?
(fittrac= yes) Fit the traced points interactively?
(extract= yes) Extract spectra?
(extras = yes) Extract sky, sigma, etc.?
(review = no) Review extractions?
(line = INDEF) Dispersion line
(nsum = 10) Number of dispersion lines to sum or median
# DEFAULT APERTURE PARAMETERS
(lower = -5.) Lower aperture limit relative to center
(upper = 5.) Upper aperture limit relative to center
(apidtab= ) Aperture ID table (optional)
# DEFAULT BACKGROUND PARAMETERS
(b_funct= chebyshev) Background function
(b_order= 1) Background function order
(b_sampl= -10:-6,6:10) Background sample regions
(b_naver= -3) Background average or median
(b_niter= 0) Background rejection iterations
(b_low_r= 3.) Background lower rejection sigma
(b_high_= 3.) Background upper rejection sigma
(b_grow = 0.) Background rejection growing radius
# APERTURE CENTERING PARAMETERS
(width = 5.) Profile centering width
(radius = 10.) Profile centering radius
(thresho= 0.) Detection threshold for profile centering
# AUTOMATIC FINDING AND ORDERING PARAMETERS
nfind = 1 Number of apertures to be found automatically
(minsep = 5.) Minimum separation between spectra
(maxsep = 1000.) Maximum separation between spectra
(order = increasing) Order of apertures
# RECENTERING PARAMETERS
(aprecen= ) Apertures for recentering calculation
(npeaks = INDEF) Select brightest peaks
(shift = yes) Use average shift instead of recentering?
# RESIZING PARAMETERS
(llimit = INDEF) Lower aperture limit relative to center
(ulimit = INDEF) Upper aperture limit relative to center
(ylevel = 0.1) Fraction of peak or intensity for automatic widt
(peak = yes) Is ylevel a fraction of the peak?
(bkg = yes) Subtract background in automatic width?
(r_grow = 0.) Grow limits by this factor
(avglimi= no) Average limits over all apertures?
# TRACING PARAMETERS
(t_nsum = 10) Number of dispersion lines to sum
(t_step = 10) Tracing step
(t_nlost= 3) Number of consecutive times profile is lost befo
(t_funct= legendre) Trace fitting function
(t_order= 5) Trace fitting function order
(t_sampl= *) Trace sample regions
(t_naver= 1) Trace average or median
(t_niter= 2) Trace rejection iterations
(t_low_r= 2.) Trace lower rejection sigma
(t_high_= 2.) Trace upper rejection sigma
(t_grow = 0.) Trace rejection growing radius
# EXTRACTION PARAMETERS
(backgro= fit) Background to subtract
(skybox = 1) Box car smoothing length for sky
(weights= none) Extraction weights (none|variance)
(pfit = fit1d) Profile fitting type (fit1d|fit2d)
(clean = no) Detect and replace bad pixels?
(saturat= INDEF) Saturation level
(readnoi= 6.1) Read out noise sigma (photons)
(gain = 1.53) Photon gain (photons/data number)
(lsigma = 4.) Lower rejection threshold
(usigma = 4.) Upper rejection threshold
(nsubaps= 1) Number of subapertures per aperture
(mode = ql)
PACKAGE = apextract
TASK = apall
input = @procarclist List of input images
(output = @specarclist) List of output spectra
(apertur= ) Apertures
(format = multispec) Extracted spectra format
(referen= @proclist) List of aperture reference images
(profile= ) List of aperture profile images
(interac= no) Run task interactively?
(find = no) Find apertures?
(recente= no) Recenter apertures?
(resize = no) Resize apertures?
(edit = no) Edit apertures?
(trace = no) Trace apertures?
(fittrac= no) Fit the traced points interactively?
(extract= yes) Extract spectra?
(extras = no) Extract sky, sigma, etc.?
(review = no) Review extractions?
(line = INDEF) Dispersion line
...
(backgro= none) Background to subtract
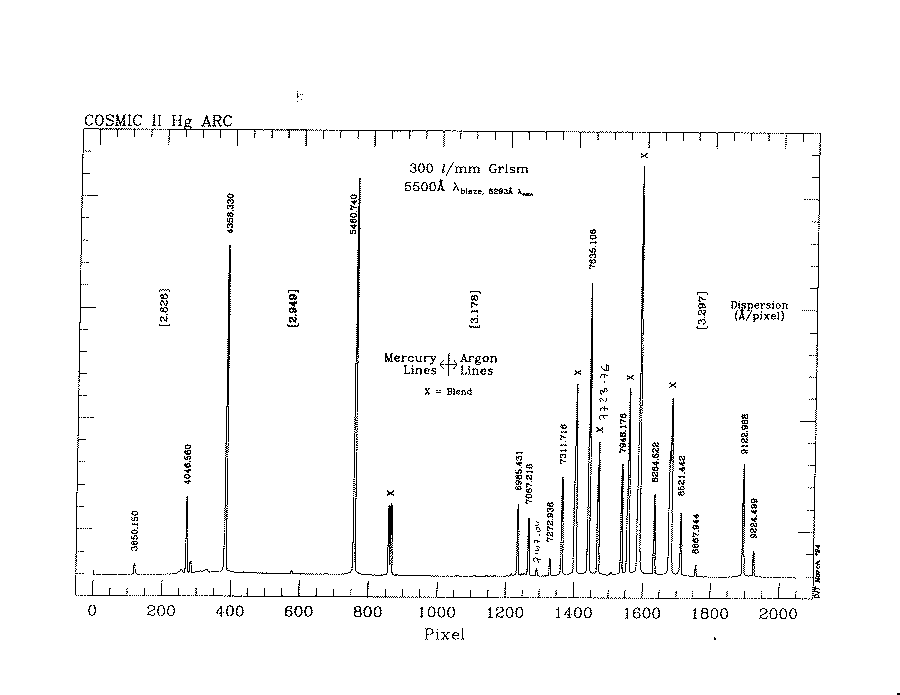
PACKAGE = onedspec TASK = identify images = specarc.29 Images containing features to be identified (section= middle line) Section to apply to two dimensional images (databas= database) Database in which to record feature data (coordli= scripts$cosmic.300lmm.arcHgAr.table) User coordinate list (units = ) Coordinate units (nsum = 10) Number of lines/columns/bands to sum in 2D image (match = 10.) Coordinate list matching limit (maxfeat= 50) Maximum number of features for automatic identif (zwidth = 100.) Zoom graph width in user units (ftype = emission) Feature type (fwidth = 4.) Feature width in pixels (cradius= 5.) Centering radius in pixels (thresho= 10.) Feature threshold for centering (minsep = 2.) Minimum pixel separation (functio= spline3) Coordinate function (order = 1) Order of coordinate function (sample = *) Coordinate sample regions (niterat= 1) Rejection iterations (low_rej= 3.) Lower rejection sigma (high_re= 3.) Upper rejection sigma (grow = 0.) Rejection growing radius (autowri= no) Automatically write to database (graphic= stdgraph) Graphics output device (cursor = ) Graphics cursor input crval = Approximate coordinate (at reference pixel) cdelt = Approximate dispersion (aidpars= ) Automatic identification algorithm parameters (mode = ql)
An example arc line map: